Intro
Before starting to write a package which helps with fuzzy matching I wanted to do a small research and explore the packages which are already out there.
I am loading {dplyr} as it is always helpful when working with data.
The data
It is very hard to come up with a realistic example which is perfect for all types of fuzzy matching algorithms. I will do a Harry Potter example and modify it along the way.
Assume that we want to find all occurrences of Harry Potter and Voldemort. As a human it is easy to see them directly, but the computer would have some problems with these examples, due to different types of spelling and typos.
input <- c("harry j potter", "harrypotter", "Voldemort",
"Harry POTTER", "Harrry Potter", "Ron Weasley")
compare <- c("Harry Potter", "Voldemort")
stringdist
The {stringdist} package by Mark van der Loo is super useful for comparing strings. And as comparison of strings is the core of the fuzzy string matching process {stringdist} is maybe the most important package to look at.
The package contains a function with the same name stringdist which calculates the distance between input and compare string. The most known method to calculate string distances is probably the Levenshtein distance which checks how many letters would have to be inserted, deleted or replaced to get from the input the the compare string.
Here, we see the distance of all input strings to the first compare string “Harry Potter”. We get six values, because our input contained six strings.
library(stringdist)
stringdist(input, compare[1], method = 'lv')
[1] 4 3 10 5 1 11You can also use the string comparison method of your choice, e.g. Jaro-Winkler. To see the whole list of available methods use the help function.
#help(`stringdist-metrics`)
stringdist(input, compare[1],
method = "jw", p = 0.1)
[1] 0.15079365 0.14393939 0.58333333 0.16666667 0.01538462 0.65909091We observe that the values are rather low if strings are similar, and higher if they are different.
So far, we have only compared the input with one of the strings (namely “Harry Potter” the first one of the compare strings).
If we would like to test all possible combinations, we can use stringdistmatrix.
stringdistmatrix(input, compare,
method = "cosine", useNames = TRUE)
Harry Potter Voldemort
harry j potter 0.13277262 0.5076340
harrypotter 0.12791840 0.4466284
Voldemort 0.46064011 0.0000000
Harry POTTER 0.38508131 0.8492443
Harrry Potter 0.01023759 0.4777670
Ron Weasley 0.62789580 0.5818790tidystringdist
If you are a fan of tidy data, you might want to have a look at Colin Fay’s {tidystringdist} which was built on top of {stringdist}. The tidy_comb function creates a dataframe with all comparisons and then tidy_stringdist calculates the distance measures.
# A tibble: 6 x 2
V1 V2
* <chr> <chr>
1 Harry Potter harry j potter
2 Voldemort harrypotter
3 Harry Potter Voldemort
4 Voldemort Harry POTTER
5 Harry Potter Harrry Potter
6 Voldemort Ron Weasley tidy_comb(input, compare) %>%
tidy_stringdist(method = c("jw", "lv", "cosine"))
# A tibble: 6 x 5
V1 V2 jw lv cosine
* <chr> <chr> <dbl> <dbl> <dbl>
1 Harry Potter harry j potter 0.151 4 0.133
2 Voldemort harrypotter 0.576 9 0.447
3 Harry Potter Voldemort 0.583 10 0.461
4 Voldemort Harry POTTER 0.602 12 0.849
5 Harry Potter Harrry Potter 0.0256 1 0.0102
6 Voldemort Ron Weasley 0.532 9 0.582 fuzzyjoin
David Robinson’s {fuzzyjoin} package is useful for so many applications. As the name already says, we are looking at joins / merges of tables here. It is the fuzzy version of left join / inner join / full outer join etc.
We will look at a small variation of our example to show how fuzzy join works. Assume that we have some extra information coming along with the compare vector, e.g. how often they use bad spells. We are working with the input, and want to add this information to our analysis. As a direct join is not possible (due to the different spellings), we can use the stringdist_join function.
library(fuzzyjoin)
df1 <- data.frame(name = c("harry j potter", "harrypotter", "Voldemort",
"Harry POTTER", "Harrry Potter", "Ron Weasley"))
df2 <- data.frame(name = c("Harry Potter", "Voldemort"),
bad_spells_index = c(0.02,0.87))
stringdist_join(df1, df2,
mode = "inner",
by = "name",
max_dist = 6)
name.x name.y bad_spells_index
1 harry j potter Harry Potter 0.02
2 harrypotter Harry Potter 0.02
3 Voldemort Voldemort 0.87
4 Harry POTTER Harry Potter 0.02
5 Harrry Potter Harry Potter 0.02Note that with larger datasets you will have to be careful with max_dist. It is the threshold of the maximal allowable string distance. If you select it too high, some names from the input will have several matches.
stringdist_join(df1, df2,
mode = "inner",
by = "name",
max_dist = 10)
name.x name.y bad_spells_index
1 harry j potter Harry Potter 0.02
2 harrypotter Harry Potter 0.02
3 harrypotter Voldemort 0.87
4 Voldemort Harry Potter 0.02
5 Voldemort Voldemort 0.87
6 Harry POTTER Harry Potter 0.02
7 Harrry Potter Harry Potter 0.02
8 Ron Weasley Voldemort 0.87And you can actually use any method from the {stringdist} package. This small paragraph is not enough to showcase the flexibility of {fuzzyjoin}. You can also do joins based on regular expressions, spatial locations and many more.
inexact
The RStudio addin {inexact} by Andrés Cruz helps to manually improve your fuzzy string matching process.
It is still under development but you can install it from Github.
remotes::install_github("arcruz0/inexact")
It helps if there are some matches which are hard to be found automatically by the program, but can be performed manually by us.
input_strings <- data.frame(name = c("Harry J. Potter","Voldemort","Lord Voldemort","Tom Riddle"),
appearances = c(50,30,20,2), stringsAsFactors = F)
compare_strings <- data.frame(name = c("Harry Potter","Voldemort","Ted Tonks"),
bad_spells = c(0.05,0.87,0.03), stringsAsFactors = F)
You can now either open in from the Addins panel on top of your Rstudio window, or run:
inexact::inexact_addin()
This opens a window which helps to fix matches manually.
knitr::include_graphics("img/inexact.gif")
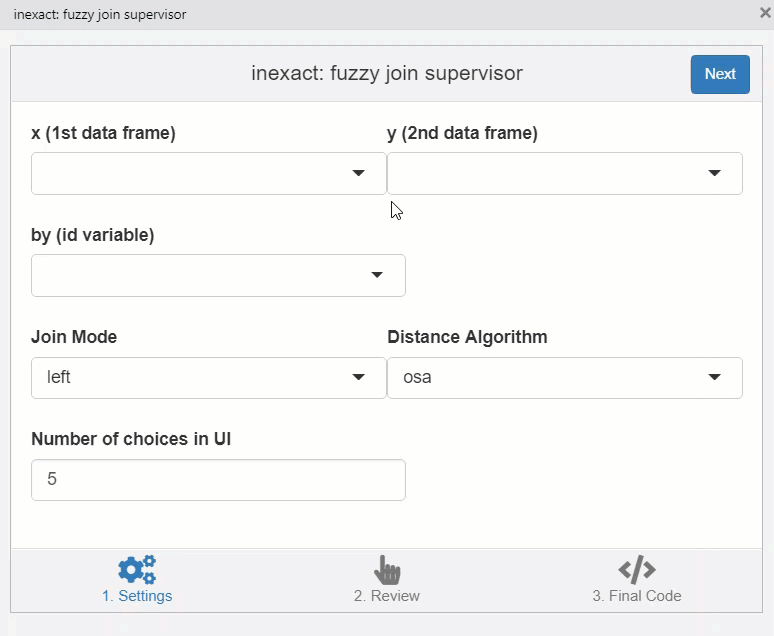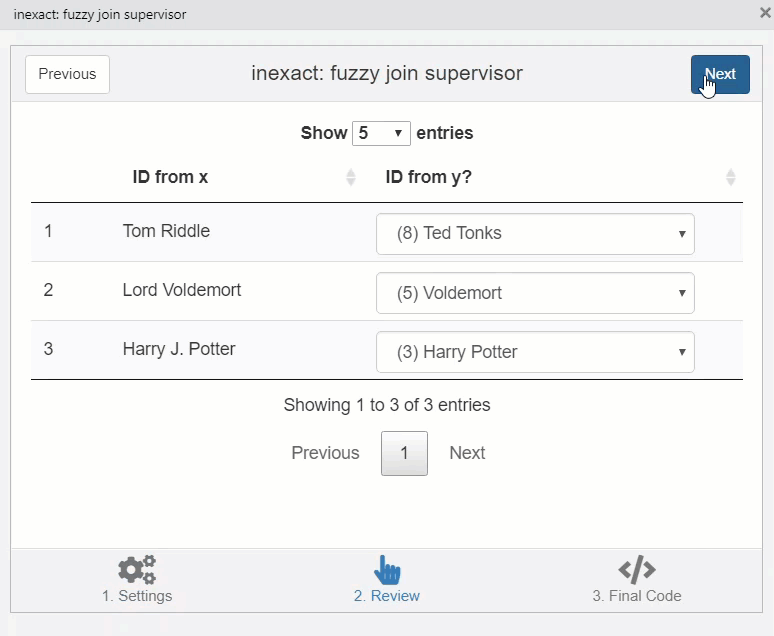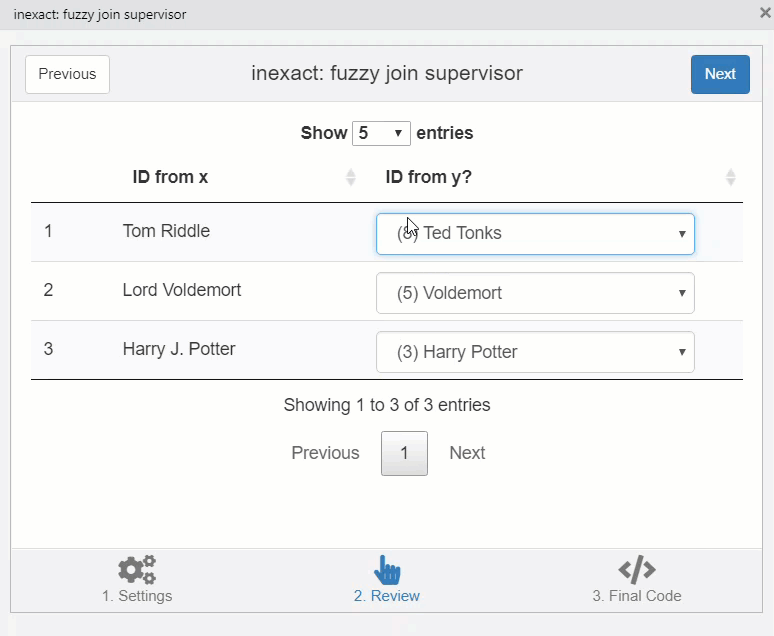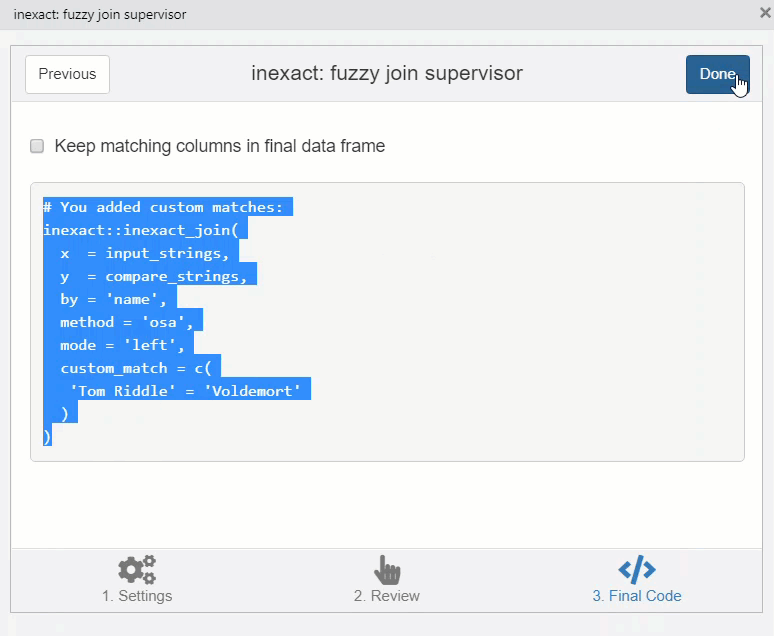
You can copy the code in the third stage of the addin to perform the join.
# You added custom matches:
inexact::inexact_join(
x = input_strings,
y = compare_strings,
by = 'name',
method = 'osa',
mode = 'left',
custom_match = c(
'Tom Riddle' = 'Voldemort'
)
)
name appearances bad_spells
1: Harry J. Potter 50 0.05
2: Voldemort 30 0.87
3: Lord Voldemort 20 0.87
4: Tom Riddle 2 0.87refinr
The {refinr} package works with string fingerprints (very good explanation of the topic here). There are different types of fingerprints, for example the 2-gram fingerprint of Harry is arharrry and the 1-gram fingerprint is ahrry.
Remember that we had one example “harry j potter”, to get the same fingerprint, we will ignore the j. I like that the compare strings are called dict, as they serve as some sort of dictionary - the ultimate truth of spelling.
library(refinr)
key_collision_merge(input, dict=compare, ignore_strings = "j")
[1] "Harry Potter" "harrypotter" "Voldemort" "Harry Potter"
[5] "Harrry Potter" "Ron Weasley" After this cleaning, we can use the n_gram_merge to replace the occurrences of Harry Potter. This method is also based on the {stringdist} package and allows for usage of other string matching methods.
input %>%
key_collision_merge(dict=compare,ignore_strings = "j") %>%
n_gram_merge()
[1] "Harry Potter" "Harry Potter" "Voldemort" "Harry Potter"
[5] "Harry Potter" "Ron Weasley" fuzzywuzzyR
When you google fuzzy string matching, you will see tons of Python articles. Most of them use the fuzzywuzzy library. The {fuzzywuzzyR} package ports this functionality to R. As far as I have seen, it only works with the Levenshtein distance.
You need to have the {reticulate} package installed which helps with the Python connection. I had some problems getting the example to work. Issues on github advice to install fuzzywuzzy and Levenshtein in Python (with pip install fuzzywuzzy) and then call reticulate::py_discover_config(required_module = 'fuzzywuzzy').
library(fuzzywuzzyR)
# Processor
init_proc <- FuzzUtils$new()
PROC <- init_proc$Full_process
# Scorer
init_scor <- FuzzMatcher$new()
SCOR <- init_scor$WRATIO #scorer function
init <- FuzzExtract$new()
init$Extract(string = "harry Potter", sequence_strings = compare,
processor = PROC, scorer = SCOR)
[[1]]
[[1]][[1]]
[1] "Harry Potter"
[[1]][[2]]
[1] 100
[[2]]
[[2]][[1]]
[1] "Voldemort"
[[2]][[2]]
[1] 29We can also try to find the best fit (or best fits - depending on the limit input) for every word.
init$ExtractBests(string = "harry j potter", sequence_strings = compare,
processor = PROC,
scorer = SCOR, limit = 1L)
[[1]]
[[1]][[1]]
[1] "Harry Potter"
[[1]][[2]]
[1] 95{fuzzywuzzyR} also has a deduplication functionality. Let’s try this with our initial input.
init$Dedupe(contains_dupes = input, threshold = 70L, scorer = SCOR)
dict_keys(['harry j potter', 'Voldemort', 'Ron Weasley'])We see no duplicates. However, the algorithm chose the first appearance “harry j potter” and adapted all the others. It would be great to have “Harry Potter” instead.
Check the vignette for more functionality and examples.
lingmatch
The {lingmatch} package does not directly work with characters but with words.
library(lingmatch)
matches <- lingmatch("Harry Potter and Voldemort are enemies", "Harry Potter does not like Voldemort")
matches$dtm
2 x 9 sparse Matrix of class "dgCMatrix"
and are does enemies harry like not potter voldemort
[1,] . . 1 . 1 1 1 1 1
[2,] 1 1 . 1 1 . . 1 1matches$sim
cosine
0.5
attr(,"time")
simets
0 There could be a potential value in fuzzy string matching if we decompose our input strings into ngrams (for example with the {quanteda} package) and use {lingmatch} with ngrams instead of words.
library(quanteda) # for the n gram decomposition
input1 <- "harry potter"
input2 <- "harry james potter"
input1_ngram <- input1 %>%
tokens("character") %>%
unlist() %>%
char_ngrams(concatenator = "") %>%
paste(collapse = " ")
input2_ngram <- input2 %>%
tokens("character") %>%
unlist() %>%
char_ngrams(concatenator = "") %>%
paste(collapse = " ")
lingmatch(input1_ngram,input2_ngram)
$dtm
2 x 16 sparse Matrix of class "dgCMatrix"
[1,] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 .
[2,] . 1 1 . 1 . . 1 1 1 1 . 1 1 . 1
$processed
1 x 16 sparse Matrix of class "dgCMatrix"
[1,] . 1 1 . 1 . . 1 1 1 1 . 1 1 . 1
$comp.type
[1] "text"
$comp
am ar er es ha ja me ot po rr ry sp te tt yj yp
1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0
$group
NULL
$sim
cosine
0.7348469
attr(,"time")
simets
0 We see that the cosine similarity of the words is 0.735.
Conclusion
There are already some great packages available to make the task of fuzzy string matching easier. It depends on the application which one or which combination of functionalities suits your needs best. As always, I am very interested in feedback and discussion: Did you already work with any of these packages? What has your experience been? What do you feel is missing to make fuzzy matching easier and better?